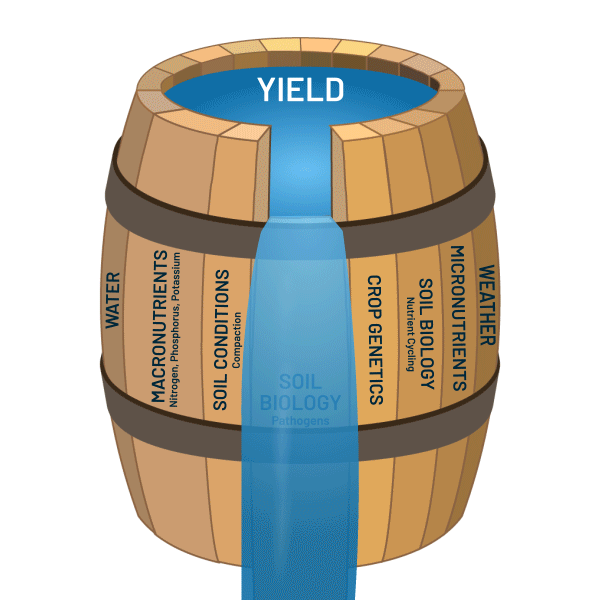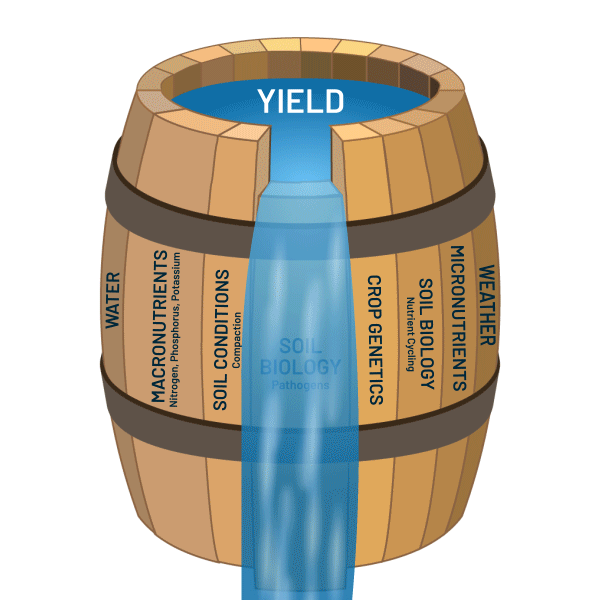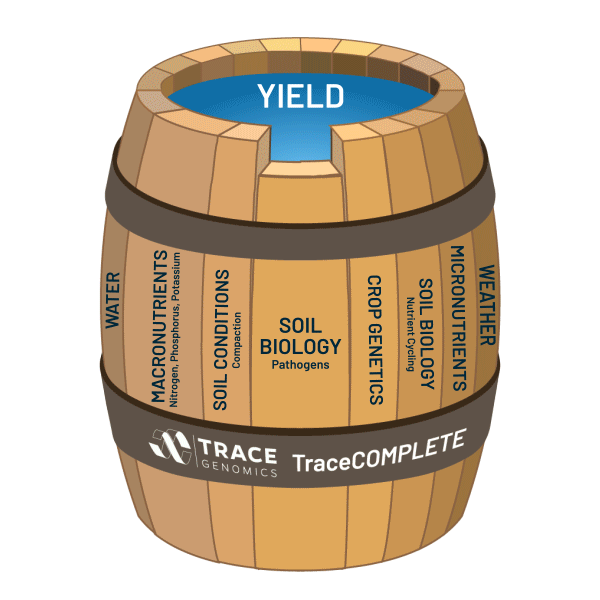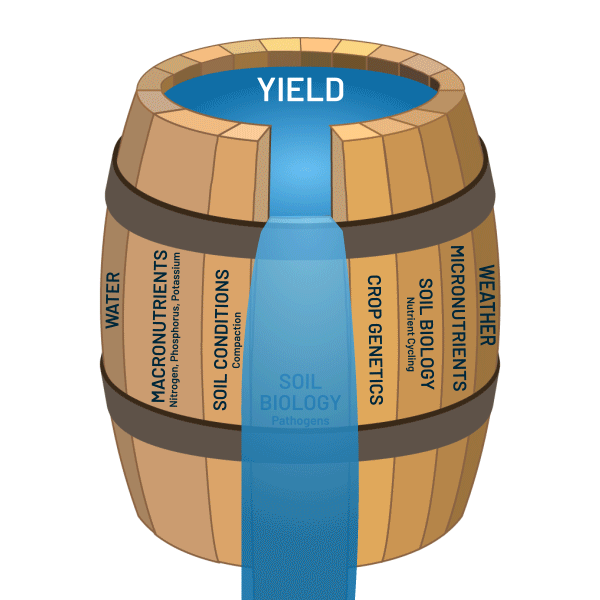
Solving Soil’s Microscopic Mysteries: Introduction to Metagenomics
At Trace Genomics, we are experts on the soil microbiome. Like your gut microbiome, the soil counterpart is composed of all the microorganisms (or “microbes”) that live there. Having this information is incredibly valuable to those who are interested in measuring soil health, including: farmers, agronomists, scientists, and any business invested in agriculture.
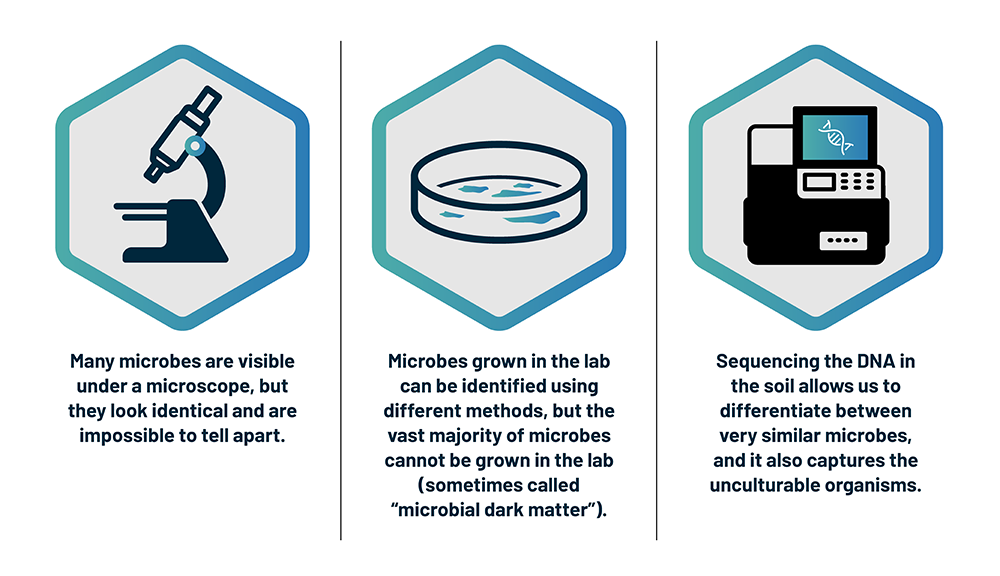
How do we know what microbes are in the soil?
There are a few different technologies that can shine a light on the invisible world of microbes.
Because it provides the most clear picture, Trace Genomics uses DNA sequencing to understand the soil microbiome. In order to understand how we do that, let’s back up a little bit…
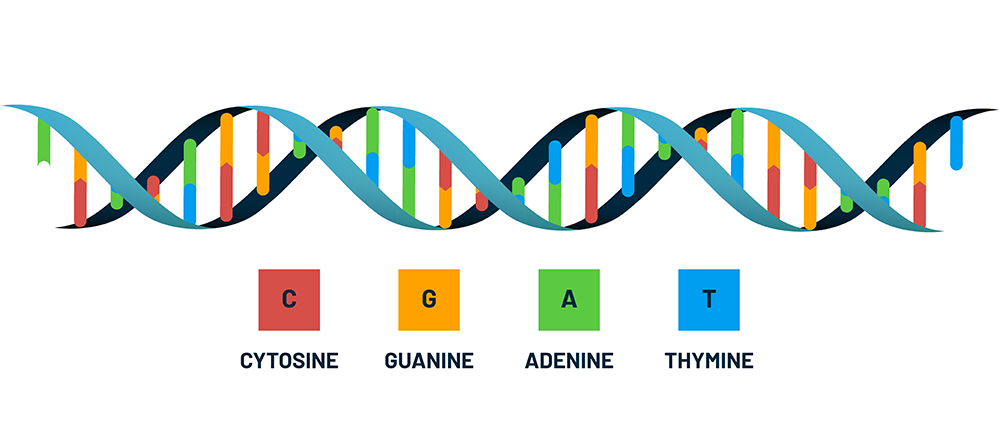
What is DNA?
DNA is known as the “blueprint of life” because it contains all the information necessary to “build” a living organism. It is a biological molecule made up of 4 different chemical building blocks called bases; you can think of it like a language with 4 letters in the alphabet.
The order of bases in a DNA molecule can be read like an instruction manual. In the manual, each chapter has instructions for a different piece of machinery (that is, another biological molecule like a protein), and the DNA that codes for it is called a gene. All the DNA for a particular organism is called its genome, and the process that scientists use to read a genome is called DNA sequencing.
Different types of environmental DNA sequencing
Metagenomics
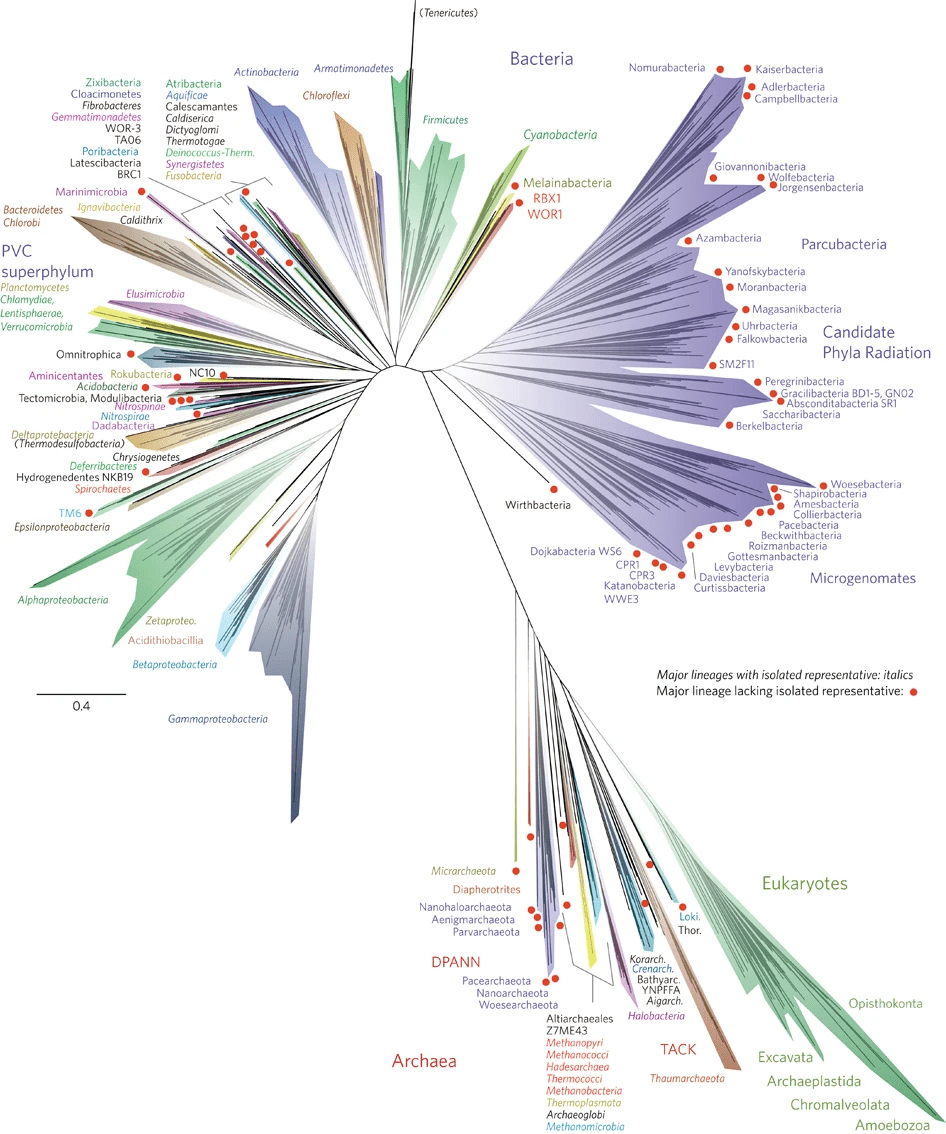
Sequencing all the DNA in an environment is known as metagenomics. In the past 20 years, this has been an invaluable scientific discovery tool for understanding the true breadth of the tree of life. Because most microbes can’t be cultured in the lab or differentiated under a microscope, we had no idea how many different species there were. Since the development of metagenomics, scientists have discovered a treasure trove of microorganisms from entire groups that were previously unknown (image 1).
Image 1. New tree of life showing major expansion (purple branch) from metagenomics. Figure 1 from Hug et al 2016.
Amplicon sequencing
Rather than sequencing all DNA in an environment, scientists can also use “fingerprint” genes to see what types of microbes are present. For each of the large groups of microbes (Bacteria, Archaea, Fungi, and Protists), there are a few genes that are found in all the members (this is called a conserved gene). For example, the 16S rRNA gene (or just “16S”) is conserved among bacteria and archaea, so scientists can sequence all instances of that gene from an environment. Importantly, while being well-conserved, the 16S gene is also variable enough to tell the difference between different groups of bacteria (though often not the difference between species).
Differences between metagenomics and amplicon sequencing
If we think of a single organism’s genome as a complete puzzle, a soil metagenome is like having 10,000 different puzzles with their pieces all mixed into the same box. Amplicon sequencing is like looking for a specific piece that all the puzzles have in common, like the top left corner, and using that to identify which puzzles (genomes) are there. Metagenomics looks at all the pieces to try and find other useful information, such as how many genomes contain certain functional genes (like nitrogen or phosphorus cycling).
| Metagenomics | Amplicon Sequencing |
|---|---|
| Sequences all DNA with one protocol | Sequences only a fingerprint gene. Different protocols are needed for bacteria, fungi, protists, etc. (and they differ in how well they work) |
| Higher resolution—can identify species/ strain level | Lower resolution—cannot reliably identify individual species. (May be able to say there is a Pseudomonas but not tell what species it is. This is problematic for groups where some members are pathogens and some are not) |
| Get direct functional information by counting functional genes (ex., in nitrogen cycling) | Hypothesize function based on the identity of microbes |
| Takes more computational power | Easier computationally |
| More expensive per sample | Cheaper per sample |
| Get valuable information even without a database (i.e., previously recorded sequences not required) | Requires database of known sequences |
About the author: Dr. Tuesday Simmons is the Science Writer at Trace Genomics. She earned her Ph.D. in Microbiology from the University of California, Berkeley, studying the root microbiome of cereal crops.