Using the Power of DNA to Improve Sustainability in Agriculture
Climate change is impacted by soil microbes, and the best way to understand soil microbes is with DNA.
What you do makes a difference, and you have to decide what kind of difference you want to make.
Climate change is one of the most serious threats to the world as we know it. Agriculture is responsible for 10% of greenhouse gas (GHG) emissions in the United States, and about half of those emissions can be attributed to soil management (1). Since Fritz Haber discovered how to turn nitrogen gas into fertilizer over 100 years ago, crop yield has skyrocketed (2). While this is good news for growing global food demand, overapplication of fertilizers also leads to leaching of nutrients into waterways, causing environmental damage in places like the Chesapeake Bay and the Gulf of Mexico. Additionally, the production and use of nitrogen fertilizers is responsible for the majority of GHG production in the soil.
With these factors in mind, growers have been working to implement more sustainable soil management practices and reduce GHG emissions. In order to develop, execute, and evaluate these practices, the soil should be considered more holistically than chemical measurements like pH and nutrients. A major, often forgotten component of soil health is the vast number of organisms that make it a thriving, living ecosystem. How vast? In 1 teaspoon of soil, there can easily be 1 billion microorganisms (3). Combined together, all the living organisms in the top six inches of a healthy soil over one acre can weigh more than 5,000 pounds (4). Understanding the diversity of species present can help us to both evaluate soil health and formulate a plan to improve it.
Many soil species are microorganisms (aka microbes) and invisible to the naked eye, so how can we know what different microbes are there? Here’s where DNA comes in. Historically, microbiologists have studied these tiny beings using microscopes or by growing them as cultures in a lab. Advances in DNA sequencing technology in recent decades have led to the development of metagenomics—sequencing all the DNA in an environment. Metagenomic analysis of soils has shone a light on the true level of biodiversity that has, until now, gone undiscovered (5). Trace Genomics has developed a groundbreaking platform for analyzing metagenomic data and translating it into tangible actions. Looking at DNA can give us both identifying information (is a pathogen present?) and functional information (can anyone here make nitrates?), all of which can be used to measure soil health.
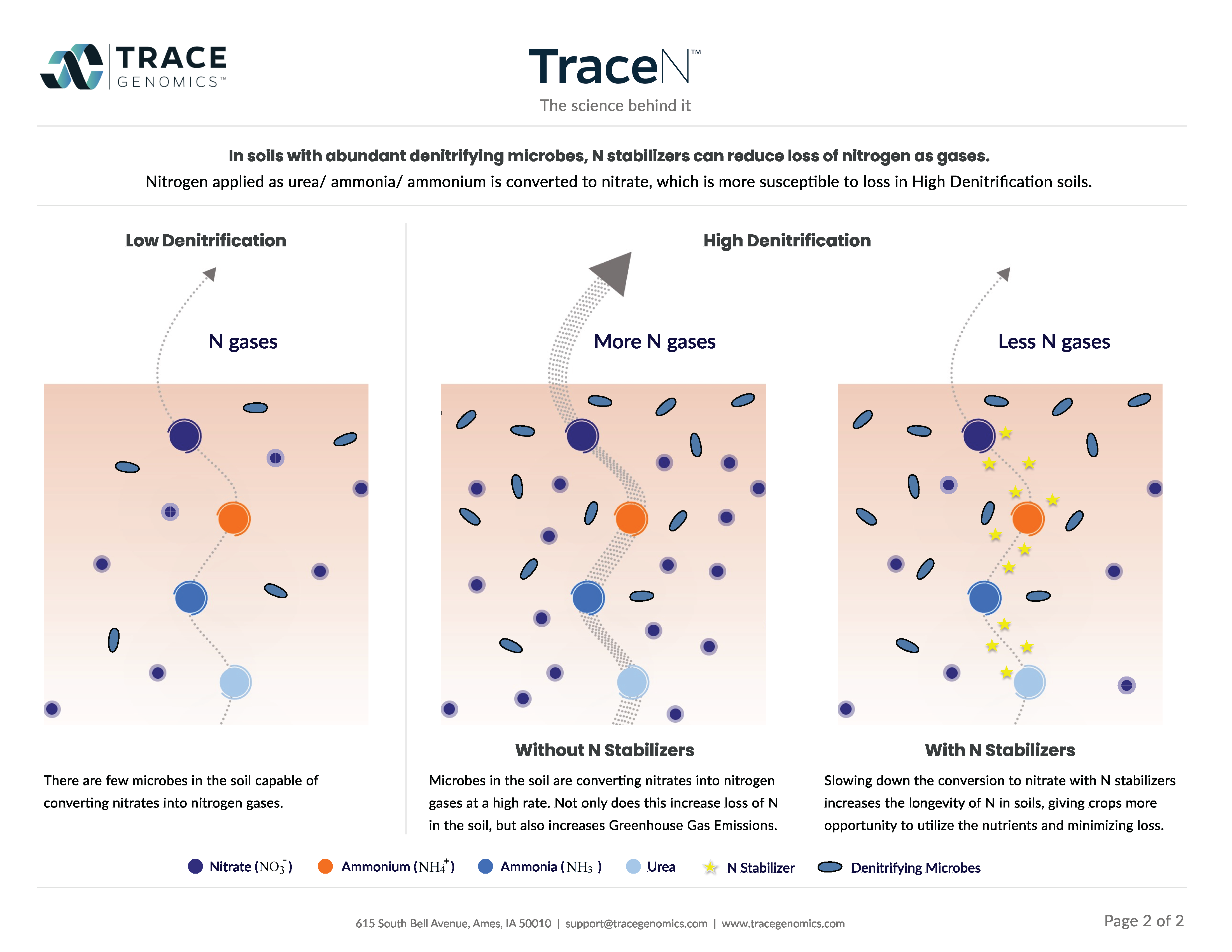
This brings us back around to climate change. With advanced technology to study soil microbes, we can better understand how they’re impacting GHG emissions. The largest and most direct way that microbes contribute to GHG levels is the production of nitrous oxide (N2O), a much more potent greenhouse gas than CO2 (6). High N2O production is more likely to occur on heavily fertilized soils (7) and those with poor drainage (8). Not all microbes produce N2O, so understanding this process isn’t as simple as measuring the amount of microbes in the soil. Trace can detect the genes responsible for N2O production in our metagenomic data, and provide growers with a benchmarked quantity of how many of these microbes are in their field.
On the flip side, microbes can also help with mitigating climate change. Microbes are able to consume the three GHG that they produce (methane, CH4, in addition to CO2 and N2O). The levels of N2O-producing and carbon sequestering microbes can be controlled through soil management practices (9, 10). With the power of metagenomics, Trace allows growers to monitor levels of these microbes, make decisions for soil management practice, and evaluate how those practices are impacting the soil microbiome.
Applying knowledge of the soil microbiome with the goal of combating climate change is not only of interest to the agronomists and growers who are directly responsible for soil management. At Trace, we also perform metagenomic analysis for two other interested parties: biological product developers and sustainable agricultural supply chain managers. Biological companies develop and manufacture products that boost plant growth using a biological rather than a chemical mechanism. These products can be live microorganisms or biomolecules produced by either microbes or plants. Due to the complexity of the soil microbiome and variables like soil chemistry and weather, it can be difficult to predict whether a biological product will be effective. Metagenomics can provide more confidence in placing these products. For sustainable supply chain managers, Trace provides a Sustainable Soil Performance (SSP) rating for soils as an objective measurement of soil health. Combining chemical and biological data on nutrients, carbon, water, and biodiversity, the SSP rating is a way to evaluate the impact of soil management practices over time. This actionable, DNA-based data leads to healthier soils and a reduction of GHG emissions.
About the author: Dr. Tuesday Simmons is the Science Writer at Trace Genomics. She earned her Ph.D. in Microbiology from the University of California, Berkeley, studying the root microbiome of cereal crops.
References
- EPA (2023) Inventory of U.S. Greenhouse Gas Emissions and Sinks: 1990-2021. U.S. Environmental Protection Agency, EPA 430-R-23-002. https://www.epa.gov/ghgemissions/inventory-us-greenhouse-gas-emissions-and- sinks-1990-2021.
- Louchheim, J. (2016, October 29). Fertilizer History: The Haber-Bosch Process. Tfi.org. https://www.tfi.org/the-feed/fertilizer-history-haber-bosch-process.
- The secret life of soil. (2010, January 8). Extension Communications. https://extension.oregonstate.edu/news/secret-life-soil.
- Healthy Soils Are: Full of Life. (n.d.). Retrieved May 25, 2023, from https://www.nrcs.usda.gov/sites/default/files/2023-01/Healthy-Soils-Are-full-of-life.pdf.
- Hug, L. A., Baker, B. J., Anantharaman, K., Brown, C. T., Probst, A. J., Castelle, C. J., Butterfield, C. N., Hernsdorf, A. W., Amano, Y., Ise, K., Suzuki, Y., Dudek, N., Relman, D. A., Finstad, K. M., Amundson, R., Thomas, B. C., & Banfield, J. F. (2016). A new view of the tree of life. Nature Microbiology, 1(5). https://doi.org/10.1038/nmicrobiol.2016.48.
- IPCC, 2013: Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change [Stocker, T.F., D. Qin, G.-K. Plattner, M. Tignor, S.K. Allen, J. Boschung, A. Nauels, Y. Xia, V. Bex and P.M. Midgley (eds.)]. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, 1535 pp.
- HOBEN, J. P., GEHL, R. J., MILLAR, N., GRACE, P. R., & ROBERTSON, G. P. (2010). Nonlinear nitrous oxide (N2O) response to nitrogen fertilizer in on-farm corn crops of the US Midwest. Global Change Biology, 17(2), 1140–1152. https://doi.org/10.1111/j.1365-2486.2010.02349.
- Lawrence, N. C., Tenesaca, C. G., VanLoocke, A., & Hall, S. J. (2021). Nitrous oxide emissions from agricultural soils challenge climate sustainability in the US Corn Belt. Proceedings of the National Academy of Sciences, 118(46). https://doi.org/10.1073/pnas.2112108118.
- SOIL MICROBES. (n.d.). KBS LTER. https://lter.kbs.msu.edu/who-we-are/research-highlights/soil-microbes/#:~:text=Microbes%20in%20terrestrial%20environments%20are.
- Bhattacharyya, S. S., Ros, G. H., Furtak, K., Iqbal, H. M. N., & Parra-Saldívar, R. (2022). Soil carbon sequestration – An interplay between soil microbial community and soil organic matter dynamics. Science of the Total Environment, 815, 152928. https://doi.org/10.1016/j.scitotenv.2022.152928.