Wide Open Spaces: Understanding Variability of Soil Biology Across Your Field
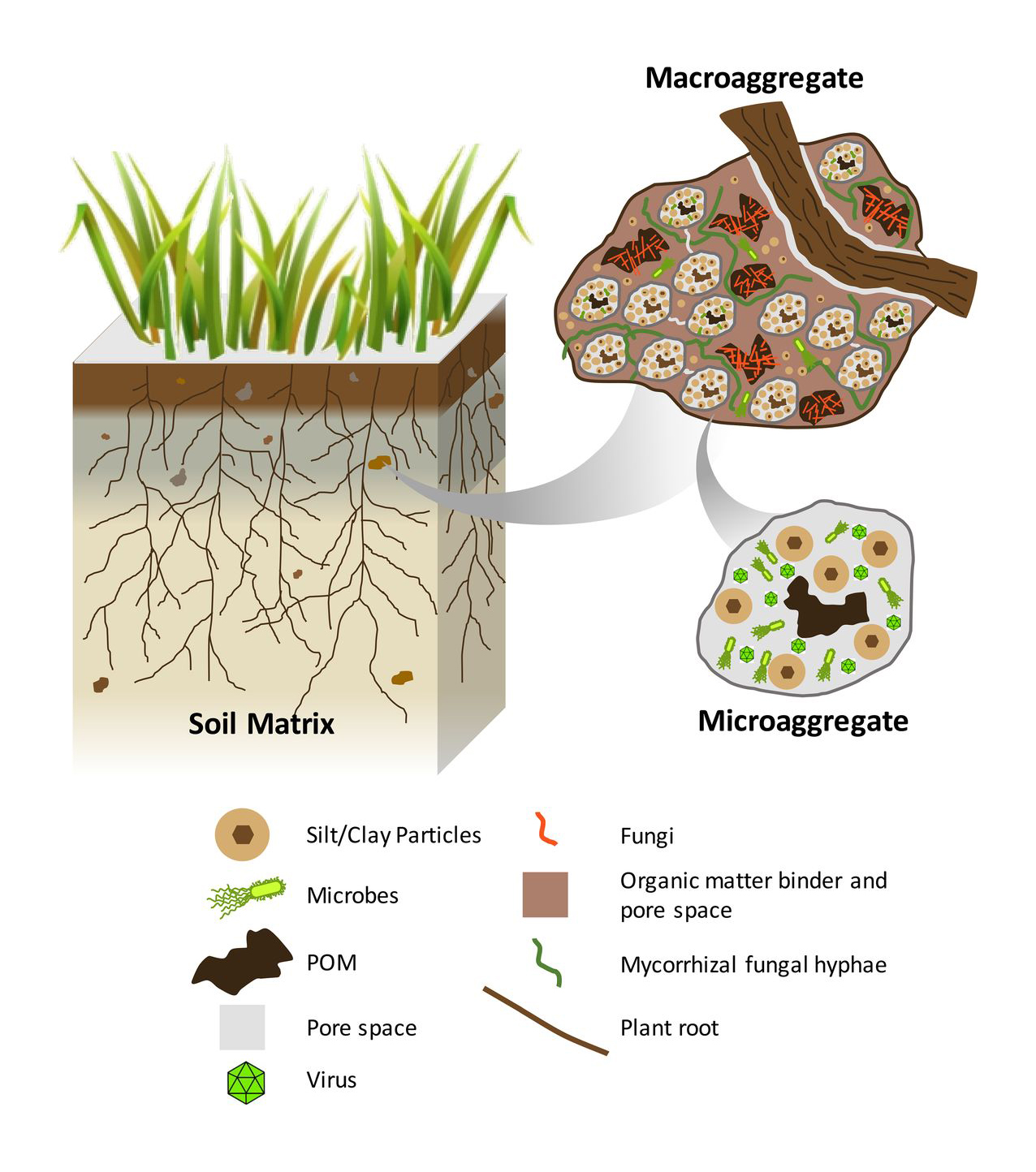
Image from Wilpiszeski et al, 2019
Soil is composed of organic and inorganic matter and has a structure more complex than what is visible by the naked eye. It contains pockets of gasses and liquids in between clumps (called aggregates) of different sizes (macro- or microaggregates). When plant roots, fungal hyphae, and subterranean animals move through the soil, they create spatial heterogeneity—tiny “islands” with unique physical and chemical characteristics.
The soil microbiome is composed of all the microscopic organisms (or microbes) living in a soil, and these are impacted by the miniscule changes in the structure and chemical composition of the soil. With so much spatial heterogeneity in soil, how are scientists able to make conclusions about the biological capacity of an entire agricultural field? A teaspoon of soil contains around a billion microbial cells, but if you pick up another teaspoon, do you see the same microbes?
How do we analyze the soil microbiome?
At Trace Genomics, our expert team of soil microbiologists have established an easy-to-follow protocol for collecting agricultural soil samples that minimizes time, effort, and cost without compromising the scientific integrity of customer samples. With TraceCOMPLETE, we use metagenomics (sequencing all the DNA in a sample) alongside our proprietary database and analytical pipeline to inform growers: 1) what microbes are living in their soil, and 2) what functional role they have. Through our online portal, TraceVIEW, agronomists can see levels of pathogens in their fields as well as microbes important in nutrient cycling. To collect soil for sequencing, we ask growers to collect three separate samples from a field. Each sample will be a composite of 4-10 soil cores (depending on depth) collected either: 1) from a radius around a central point, or 2) in a zigzag pattern throughout a plot.
How many samples are necessary for a single field?
Considering the large area of agricultural plots and the heterogeneity of soil, it is reasonable to initially expect that three samples would not be enough to draw conclusions about the microbiome function of a plot. However, due to the homogeneous treatment of a plot, there is not as much variation as you may anticipate. The most important factors that determine what microbes can live in a soil are: pH, moisture, nutrient levels, organic matter, temperature, and the plant species growing there (1). If a farmed area has a consistent application of inputs (nutrients, pesticides, etc.) and an evenly distributed monoculture crop, these factors do not vary significantly across a field. Since the microbiome is controlled by these soil characteristics and not geography (2, 3), we don’t need a large number of samples from the same field.
For perennial crops grown in orchards or vineyards, the sampling location is very important; we know the microbiome within the root zone will differ from grassy aisles between rows. However, most microbiome variation is found between plots with different soil treatments or varietals rather than within a single plot (4). Scientific studies have found that there is some microbiome variation on a smaller scale, but composite samples made up of multiple soil cores are sufficient for comparison between different plots (5). Multiple samples will result in a more accurate measurement of biodiversity, so we ask for three samples. To prevent missing a pathogen due to uneven pathogen spread, we ask growers to select samples in areas of high, medium, and low productivity or in areas where they have inconsistent or unexplained yields. Increasing the number of samples from three can reveal rare soil microbes, however from a practical standpoint, rare microbes will not have an impact on crop growth or nutrient cycling.
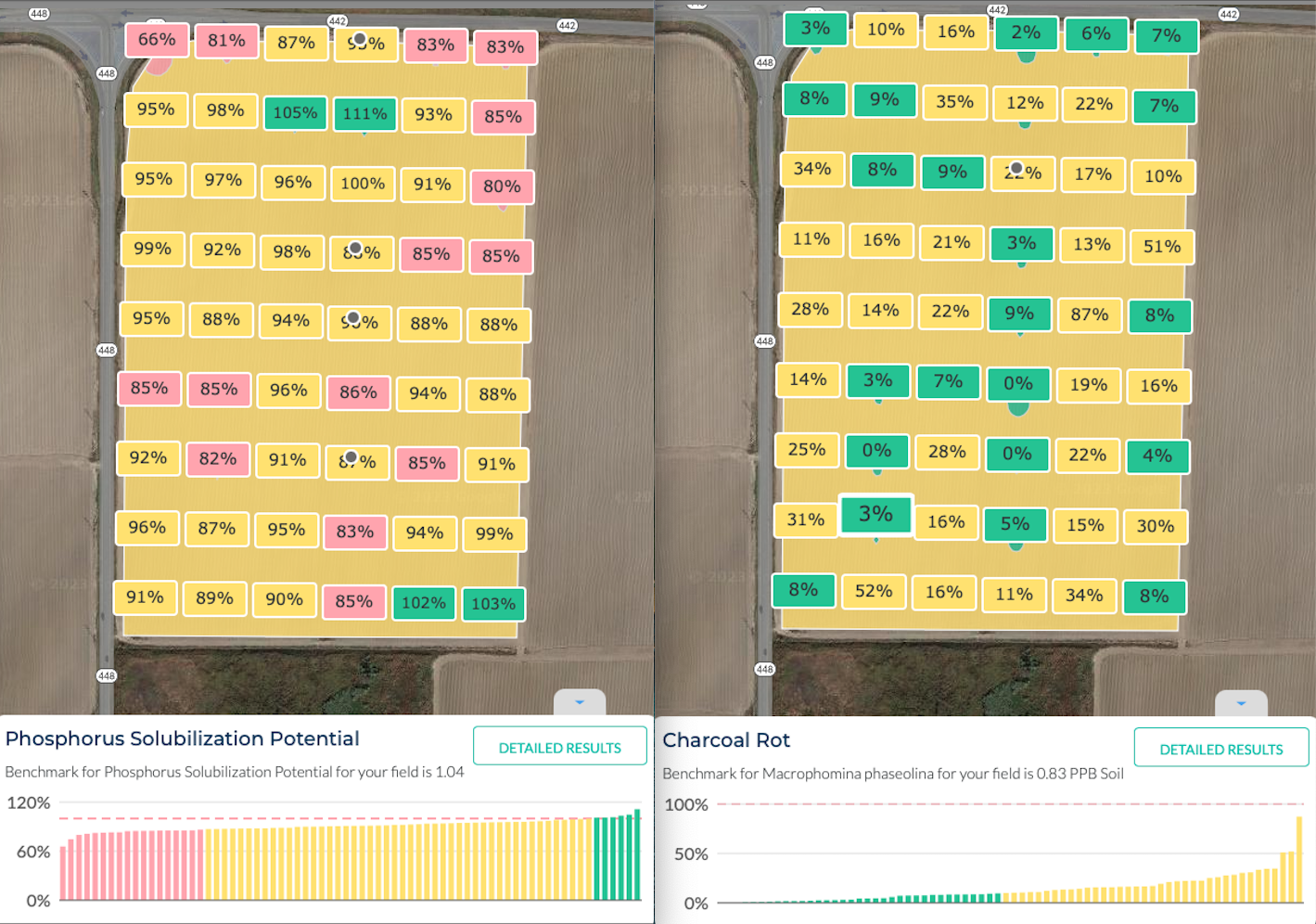
Data from the field
In the data presented below, we used TraceCOMPLETE to look at soil samples from 1-acre grids across a large field. We see that there is very little variation across a field for nutrient cycling indicators (left panel below). Pathogens may be more variable with hotspots, but we found that they are still consistent across a field (right panel below).
Conclusion
Despite micro-scale heterogeneity of soils, the large-scale homogeneous treatment of agricultural fields makes the driving factors of soil microbiome variation consistent across a large area. For this reason, three samples composed of multiple soil cores can tell a nearly complete story about a soil’s biological potential.
About the author: Dr. Tuesday Simmons is the Science Writer at Trace Genomics. She earned her Ph.D. in Microbiology from the University of California, Berkeley, studying the root microbiome of cereal crops.
References:
- Custódio, V., Gonin, M., Stabl, G., Bakhoum, N., Oliveira, M. M., Gutjahr, C., & Castrillo, G. (2021). Sculpting the soil microbiota. The Plant Journal, 109(3), 508–522. https://doi.org/10.1111/tpj.15568
- Fierer, N., & Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences, 103(3), 626–631. https://doi.org/10.1073/pnas.0507535103
- Lee, S. A., Kim, J. M., Kim, Y., Joa, J. H., Kang, S. S., Ahn, J. H., Kim, M., Song, J., & Weon, H. Y. (2020). Different types of agricultural land use drive distinct soil bacterial communities. Scientific Reports, 10(1). https://doi.org/10.1038/s41598-020-74193-8
- Deakin, G., Tilston, E. L., Bennett, J., Passey, T., Harrison, N., Fernández-Fernández, F., & Xu, X. (2018). Spatial structuring of soil microbial communities in commercial apple orchards. Applied Soil Ecology, 130, 1–12. https://doi.org/10.1016/j.apsoil.2018.05.015
- Castle, S. C., Samac, D. A., Sadowsky, M. J., Rosen, C. J., Gutknecht, J. L. M., & Kinkel, L. L. (2019). Impacts of Sampling Design on Estimates of Microbial Community Diversity and Composition in Agricultural Soils. Microbial Ecology, 78(3), 753–763. https://doi.org/10.1007/s00248-019-01318-6